🦉🌵🌎💻 Informatics to the Rescue of Biology: Global Biodiversity Information Facility (Part 2: GBIF API)
🦉🌵🌎💻 Informatics to the Rescue of Biology: Global Biodiversity Information Facility (Part 2: GBIF API)
By Enio...
Hello, friends of the great multidisciplinary community #SteemSTEM and readers from all over the world. Continuing with the topic of Global Biodiversity Information Facility (GBIF), this time I will emphasize an important and attractive aspect for computer scientists and programmers who wish to contribute to biodiversity research or the development of related applications: the GBIF API.
It has been seen that this service is the largest information system in the world, created to facilitate access to a wide range of data on biodiversity around the planet, being fed by more than a thousand institutions and used by thousands of others. Its mission has to do basically with the promotion of scientific research, and after this, many ideas that derive in initiatives that go from the conservation of species to look for ways to generate "economic benefits". [1][2]
The system's official website is hosted after the URL https://www.gbif.org/, which is its main frontend. It has also been said that it has a software called Integrated Publishing Toolkit IPT, which is the means by which affiliated institutions grow and maintain the network of repositories and available data. [3]
About GBIF API and Client Libraries
Now, other great features that I wanted to examine of the GBIF has to do with programming, a more technical component of the system. The organization after the GBIF has made available a complete Application Programming Interface of type RESTful with responses in JSON format, thought for the automation of the use of the GBIF through applications developed by third parties.
The idea of an API is to provide a more sophisticated and flexible access point to the database, which makes the existing information available in a way that is easily usable and manipulable through programming techniques. On the API can be built third party services (commercial or not) or applications that automate many tasks related to that information.
According to official documentation, the GBIF API is divided into "logical sections" (methods) for easy manipulation. Image 1 contains a table with the main methods covered by the API (see image 1). [4]
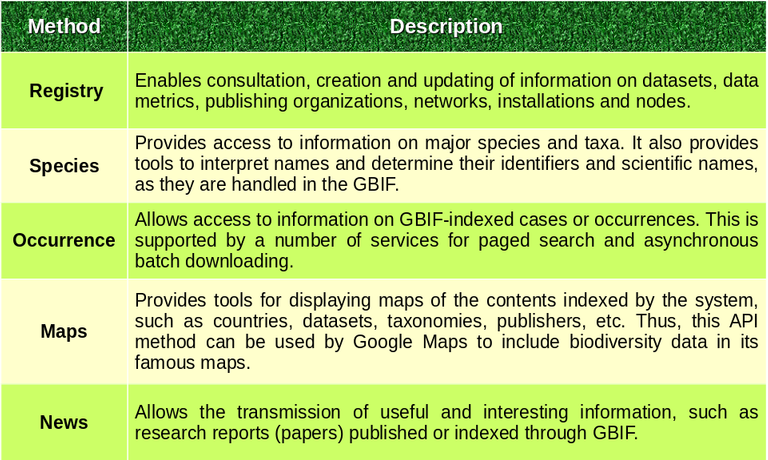
⬆️ Image 1: methods supported by the GBIF API. Author: Eniolw based on official API information [4] License: public domain
However, programmers usually do not interact with the API directly, but do so through libraries or wrappers, which are prefabricated pieces of software that join the project under development and act as an intermediary with the API, greatly facilitating the programming work.
There are three known libraries to work with GBIF, developed by Professors Scott Chamberlain and Carl Boettiger, both from University of California. These libraries are: rgbif, written in R language; pygbif, written in Python language; and gbifrb, written in Ruby language.
The review of the source code of each library, together with the details about the documentation provided, point out that not all libraries have the same degree of development or functionality. The most advanced is the rgbif library, which in the context of the R language is called 'software package'. The R language is very popular in academic and scientific environments, and greatly facilitates the manipulation, statistical processing and visualization of data. Perhaps this is why it was selected to create the most complex of libraries.
On the other hand, the least sophisticated library is gbifrb, which works in the Ruby language environment. Although this language is not very common in scientific environments, it is in more general environments, especially at the level of web services. That could justify why this library was created for Ruby, in order to give greater scope to the API of GBIF, allowing the proliferation of all types of web application client.
Finally, there is the library pygbif, written in the Python programming language. This language is as popular in academia and science as it is in other types of contexts (desktop, multiplatform software development, scripting, web development, graphical interfaces, videogames, Big Data, etc.), because it is a general purpose language quite widespread, developed and documented. For this reason, the revision of the library for Python will be done in detail.
In this sense, pygbif is distributed as free software and open source under the MIT license, with public repository in GitHub [8]. It can be easily installed from the Python Packet Indexer (PyPi) with the command: pip install pygbif. To date, the GBIF API methods supported by the library as modules are: registry, species and ocurrences. [5]
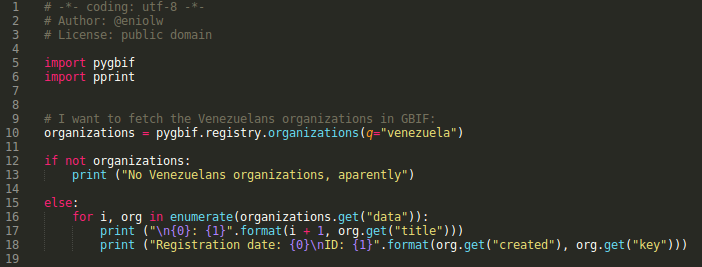
The registry module contains information about datasets, nodes, installations, networks and organizations related to GBIF. Next, a demonstration script created using this module is inserted (see image 2).
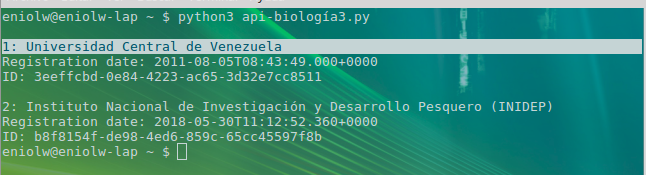
In this script there is basically a query to know which records of organizations have the word "Venezuela", which helps to locate those organizations in this country that are linked to GBIF. Specifically, it is interesting to obtain and show the title of the organization, data about the time it was registered (date, time) and its identification code (ID). The output of the script execution is presented below (see image 3).
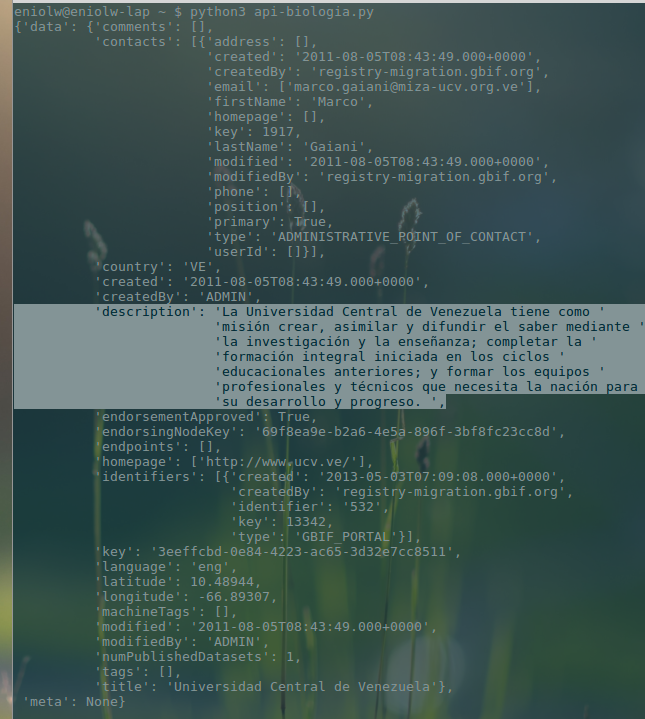
As can be seen, there are two registered Venezuelan organizations: Universidad Central de Venezuela (UCV) and Instituto Nacional de Investigación y Desarrollo Pesquero (INIDEP). To illustrate the usefulness of the ID field of the registries, a new query can be made using the ID of the Central University of Venezuela as follows:
pygbif.registry.organizations(uuid="3eeffcbd-0e84-4223-ac65-3d32e7cc8511")What a 'pure', unprocessed output like the following would produce, where the description attribute has been highlighted to confirm that this is indeed the Venezuelan university (see image 4).
In turn, the species module concentrates on handling taxonomic names. It should be remembered that taxonomy refers to the classification of organisms and this classification is quite complex. The module does its work through a series of search functions, which makes it easier to find the exact names used by the GBIF, as if they were a standard, since there are many organizations with their own names and taxonomic keys. This also helps when implementing mechanisms that use URLs or the like, so that a name is always associated with a particular result.
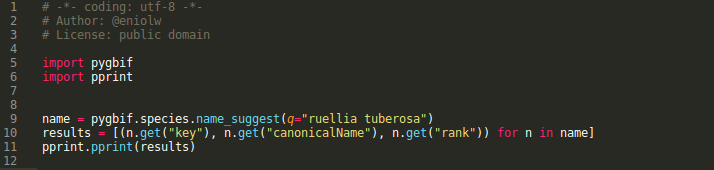
The following demonstration script makes a query using the function name_suggest, which returns 'quick' results that are used to make other decisions in the programs that are designed (see image 5).
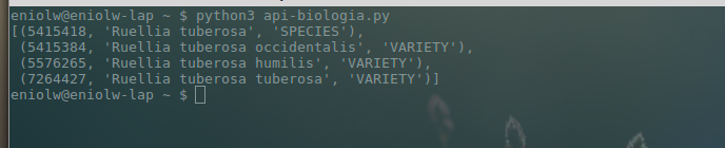
It is observed that the name ruellia tuberosa has been searched (line 9) and the values are extracted: key, canonical name and rank, to know at what height of the taxonomy the name is found (line 10). The following image shows the results (see image 6).
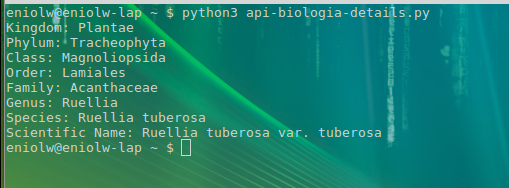
The query returned four results, one of which corresponds to the 'species' and three to 'varieties' or populations of the same species. Now, the canonical name of the last result (Ruellia tuberosa tuberosa) is used to make a more exhaustive second search with the function name_backbone. This can be seen in the following script and its corresponding output (see images 7 and 8).
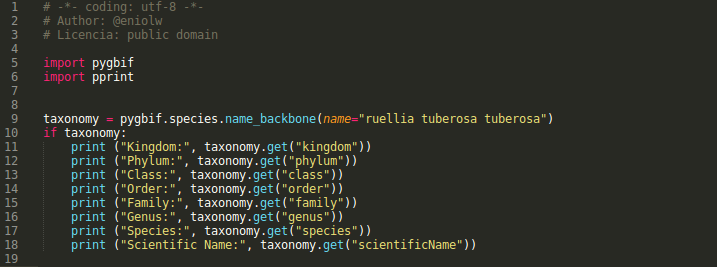
In this last script, basically, the complete taxonomy of the given variety is consulted and the specific ranges or categories of the taxonomic tree are selected (kingdom, phylum, class, order, family, genus and species). This can surely be useful enough to develop many types of applications, for example, some that verify the taxonomy of the names present in an internet publication or simply as a support tool for the researcher.
Finally, the module occurrences is commented, with which the 'cases' or information units of specimens of species are accessed, which includes a lot of interesting information, such as their contextual description as a result of research, their geolocation, associated multimedia (images), etc.
The following script is a prototype that demonstrates the potential use of this occurrence service provided by the GBIF API (see images 8 and 9).
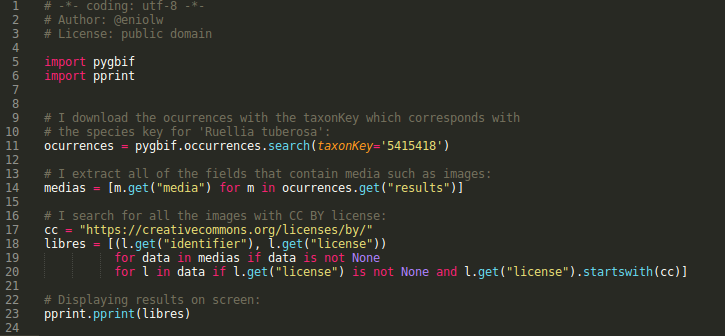
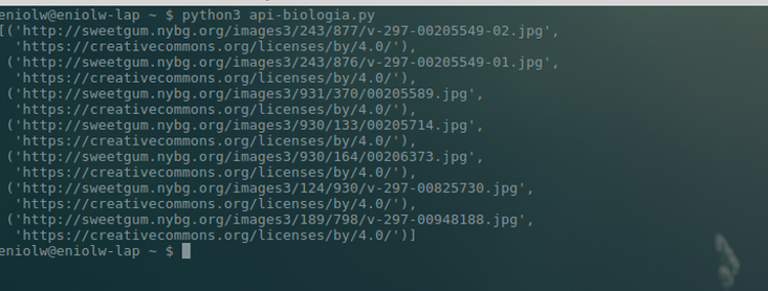
⬆️ Images 8 and 9: example of use of the occurrence module (script + list of photos URLs). Author: @Eniolw License: CC BY 3.0
As commented in the code (image 8), the idea is to extract URLs of the images associated with the occurrences of a certain species (which in the example is Ruellia tuberosa). The URLs are extracted from those images whose licenses are 'Creative Common BY', that is: free as long as attribution is given to the author. With this, seven results were obtained organized as tuples with the URL of the image and the URL to the license (image 9).
It should be noted that when opening these links, the images have a note of ' copyright reserved' included, which calls attention considering that the metadata returned by the API indicates that the license of these images is CC BY 4.0. Due to this apparent contradiction, the images referred to in the query are not published in this article, but a photograph of the same species is provided as an example (see image 10).

⬆️ Image 10: Ruellia tuberosa Author: @lupafilotaxia (published with the consent of the author)Available: here
Summary
Biodiversity is the different forms of life that inhabit the planet. The human being in his prodigious capacity to study everything, has developed important knowledge to deepen his understanding of the natural world, from the progressive accumulation and compilation of an extensive amount of information about the surrounding life. This could explain why the conversationalist Thomas Lovejoy commented that "natural species are the library from which genetic engineers can work".
This inductivist formula is powered today by computational power, in terms of information technologies that ensure the recording, manipulation and dissemination of data on biodiversity, and one of the great examples of this has been precisely the Global Biodiversity Information System (GBIF), which provides the largest network of repositories of free scientific information on biodiversity in the world.
The GBIF has some key tools to be accessible to life science researchers, who have the quite practical website and the Integrated Publishing Toolkit (IPT), a client to be exploited by affiliated organizations, such as universities, research institutes, etc. Similarly, computer scientists and programmers who contribute to biodiversity research or develop computer applications can use the GBIF API to have 'repowered' access to the resources offered by the system.
For the latter, several free, libre and open source software libraries are available, such as rgbif,pygbif and gbifrb, of which pygbif was discussed and applied in several demonstration programs during this article. It should be noted that the pygbif library can be quite satisfactory, although it is the rgbif library the most advanced of those considered, being able even to render images with the distribution and geolocation of the species (maps), all of which is a potential topic for future projects.
REFERENCES AND RESOURCES USED
- [1] GBIF (n/f). What is GBIF. Official website document from GBIF.
- [2] GBIF (2010). Memorandum of Understanding for the Global Biodiversity Information System. Document consulted here.
- [3] GBIF Secretariat (2017). Introduction to publishing using the GBIF Integrated Publishing Toolkit (IPT). Tutorial available here
- [4] GBIF (s/f). API Summary (API documentation for developers). Available here
- [5] Chamberlain, S. (2016). Pygbif library. Public repository in GitHub.
If you are interested in more Science, Technology, Engineering, and Mathematics (STEM) topics, check out the #SteemSTEM tags, where you can find more quality content and also make your contributions. You can join the #SteemSTEM Discord server to participate even more in our community and consult the periodically reports published by @SteemSTEM.
EXPLANATORY NOTES
- Unless otherwise indicated, the images in this publication have been produced by the author, including the initial image, created with images in the public domain.
Very cool Post again. Can't wait to have some time for trying this by myself. Enjoy your Weekend. Chapper
Posted using Partiko Android
Thank you very much, @chappertron!
This post has been voted on by the SteemSTEM curation team and voting trail. It is elligible for support from @curie and @utopian-io.
If you appreciate the work we are doing, then consider supporting our witness stem.witness. Additional witness support to the curie witness and utopian-io witness would be appreciated as well.
For additional information please join us on the SteemSTEM discord and to get to know the rest of the community!
Thanks for having added @steemstem as a beneficiary to your post. This granted you a stronger support from SteemSTEM.
Thanks for having used the steemstem.io app. You got a stronger support!
Thanks!
Hi @eniolw!
Your post was upvoted by Utopian.io in cooperation with @steemstem - supporting knowledge, innovation and technological advancement on the Steem Blockchain.
Contribute to Open Source with utopian.io
Learn how to contribute on our website and join the new open source economy.
Want to chat? Join the Utopian Community on Discord https://discord.gg/h52nFrV
Awesome! thanks, Utopian.