“Identify the molecule" contest #1 [Lysozyme]. Discussion. Winner announcement
In this post I’ll announce the winner and discuss the first riddle of “Identify the molecule” contest.
Thanks to all participants and everyone who showed interest and support to "Identify the molecule" contest.
(for more information go to Identify the molecule contest. Introduction), and congratulations to @scienceblocks for solving the first riddle (Lysozyme) and getting 100 STEM.
See screenshot at the end of the post for proof or go to https://stemgeeks.net/@alexbiojs/transfers
Riddle discussion.
Lysozyme was discovered accidentally near 100 years ago by Sir Alexander Fleming (the scientist known for his discovery of penicillin (1928)) in 1921. It happened accidentally, when a drop from his nose fell onto an agar plate with some microbes. He concluded that the drop contained a lytic substance, when he saw that the microbes were fading. Thus, he discovered antimicrobial properties of lysozyme.
It was the first antibiotic discovered [1].
Discovery of lysozyme was so important, that there medals and sculpture/s dedicated to this molecule.
Sculpture
"Tears" sculpture was created by Mike Tyka in 2015.
Bronze part represents the carbohydrates of the bacteria cell wall. Lead glass part represents lysozyme [4].

Discovery of the atom structure led to the first detailed enzymatic mechanism to be described and was a major breakthrough in our understanding of how our bodies' metabolic functions
[4].
The Fleming-Lysozyme Medal

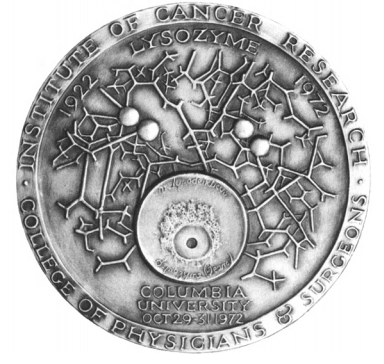
(Images were taken from "Lysozyme." book [3])
This medal was created to commemorate the fiftieth anniversary of the discovery of lysozyme by Sir Alexander Fleming and to mark the occasion of the Lysozyme Conference held at Arden House, Harriman, New York,
October 29-31, 1972.
On the obverse side of the medal we can see Sir Alexander Fleming.
On the right of this side are M. lysodeikticus (known today as Micrococcus luteus) (untreated bacteria (upper field) and treated with lysozyme bacteria (lower field)).
On the left we can see "Chance favors the prepared mind" dictum (given initially by Louis Pasteur).
On the reverse side of the medal is a culture plate with M. lysodeikticus. At the centre of it is a zone with Fleming‘s tears (which led to bacterial lysis).
The labeling of "M. lysodeikticus" and "lysozyme (tears)" is in Fleming's own hand.
[source - [3]]
Above that plate you can see part (active site) of the 3-D structure of hen egg-white lysozyme.
as defined by the X-ray crystallographic studies of David Phillips and his associates. The four circles represent the oxygens of the aspartic 52 and glutamic 35 residues which specifically accomplish the hydrolytic cleavage of the lysozyme substrate.
[source - [3]]
The medal was created by Abram Belskie [3].
Lysozyme is an enzyme, also known as muramidase or N-acetylmuramide glycanhydrolase, because it helps to break (1->4)-beta-linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues in peptidoglycans. [9]
In other words, lysozyme helps to break the cell wall of bacteria, which leads to their death.
Among other names of this molecule are N,O-diacetylmuramidase, L-7001, PR1-lysozyme, globulin G, globulin G1, mucopeptide N-acetylmuramoylhydrolase, mucopeptide glucohydrolase, muramidase [12].
Lysozyme belongs to glycoside hydrolases class of enzymes. In a nutshell, it means that it is an example of enzymes which help to break complex sugars by using water molecules.
It has been shown that the key amino acids responsible for enzymatic activity of lysozyme in chicken egg white are glutamic acid 35 (Glu35) and aspartate 52 (Asp52) (where the number represents the position of amino acid in the polypeptide chain) (remember that medal above). It’s this pair of amino acids, which uses the water molecules to split the sugar [14].
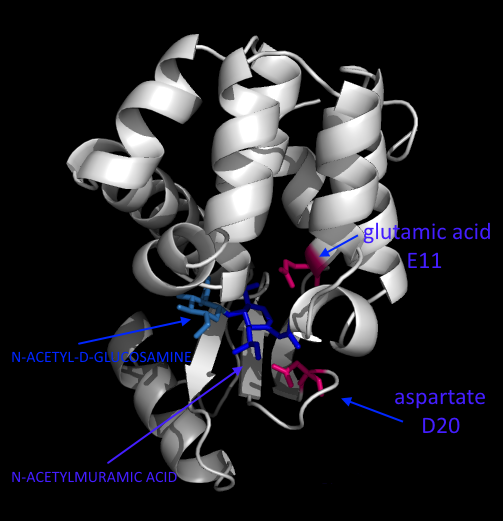
What you see above is the lysozyme of Escherichia virus T4 (that bacteriophage which infects Escherichia coli).
Analogues residues to glutamic acid 35 (Glu35) and aspartate 52 (Asp52) of egg white in this case are glutamic acid 11 (E11) and aspartate 20 (D20) (highlighted in hotpink). They help to break the linkage between N-acetylmuramic acid (highlighted in blue) and N-acetyl-D-glucosamine (highlighted in sky blue) [13].
They suppose that the origin of lysozyme goes back approximately 400 to 600 million years [15].
Lysozyme is the first protein, which was found to have all the 20 usual amino acids, it is the first enzyme to have its mechanism of action revealed, and it’s the first enzyme for which scientists solved its 3D-structure (with the help of X-ray diffraction methods). It’s a small and stable enzyme.
All that makes lysozyme an ideal candidate for research of protein structure / function. That’s why Brian Matthews (University of Oregon) used this protein to study functional changes of proteins due to structural changes (creating new active sites, removing large residues inside the protein etc.). He created so many mutants of lysozyme, that it became the most common protein in the PDB [8].
In spite of that, not everything is understood about this molecule.
It turned out that antibacterial properties of lysozyme are due to not only its enzymatic activity, but also non-enzymatic actions / structural factors [10].
Lysozyme is a promising “tool” to increase our chances to fight malaria.
The result of treatment highly depends on early and accurate
diagnosis.
In particular, it is essential to diagnose the presence of P. falciparum early since the infection can be fatal within few days.
[source - [2]]
And lysozyme might serve as a biomarker for disease severity [2].
It ca be found not only in animals, but also in plants, bacteria and bacteriophages (the latter use it to break into the host bacterial cell to inject their DNA).
Lysozyme can be found in all animal tissues, fluids and in most of the secretions [2].
It’s an important player of our innate immune system with anti-bacterial/fungal/viral properties [2].
Respiratory and gastrointestinal tracts are some of the most important anatomical barriers to microbes [11]. That’s why (I suppose) we see that the gene of lysozyme is expressed the most in trachea, bronchus, tongue tissue, stomach tissues etc.

References
Lysozymes: model enzymes in biochemistry and biology. P. Jollès (Ed.). Birkhaüser Verlag, Basel. viii + 469 pages, SFr198 (1996).
Human and Mosquito Lysozymes: Old Molecules for New Approaches Against Malaria. Mauro Prato, 2015th Edition.
Lysozyme. ELLIOTT F. OSSERMAN, ROBERT E. CANFIELD and SHERMAN BEYCHOK, 1974.
Important
This contest is the work in progress.
This is just the beginning.
A lot can be changed in the future.
Any recommendations/suggestions are very welcome.
All images (without the license specified) are used under the doctrine known in USA as "Fair Use" (similar doctrines are used in other countries). For more information visit the US Gov website